INTRODUCTION
In 2016, the Third International Consensus Definitions for Sepsis defined sepsis as “a life-threatening organ dysfunction caused by a dysregulated host response to infection”.[1,2] Sepsis is a serious acute inflammatory condition with high long-term mortality and a reduced life span of less than one year after admission.[3] While sepsis can be caused by bacteria, fungi, viruses, or parasites,[4] this study focused specifically on bacterial sepsis because of its prevalence and the significant burden placed on healthcare systems.[5] Bacterial infections are the leading cause of sepsis globally, and bacterial sepsis has distinct pathophysiological features that warrant focused discussion.[6] Additionally, many recent diagnostic and therapeutic advancements have concentrated on bacterial sepsis, making it a relevant and timely subject for review.
Sepsis remains one of the primary causes of mortality in intensive care units (ICUs) in developed nations.[7] In middle- and low-income countries, malnutrition and a lack of access to antibacterial vaccines are among the factors that contribute to the alarming increase in the incidence of bacterial sepsis. Although the number of published studies addressing the inflammatory, immunosuppressive, and procoagulant aspects of sepsis have increased over the past two decades, the latest statistics revealed that mortality because of sepsis has not improved despite a significant increase in pathomechanistic understanding. According to a recent scientific report, approximately 48.9 million cases of sepsis and 11 million sepsis-related deaths occurred globally in 2017, accounting for nearly 20% of all worldwide fatalities. The study also revealed that nearly half of all reported sepsis cases worldwide in 2017 occurred in children, with an estimated 20 million cases and 2.9 million deaths in children aged under 5 years.[8]
The emergence of drug resistance is of global importance in the management of sepsis. Early diagnosis and appropriate treatment of sepsis are vital for increasing patient survival.[9] Unlike other life-threatening conditions, such as cancer and myocardial infarction, there is no gold standard diagnostic test. Biomedical scientists face several key challenges when developing accurate and rapid tests for diagnosing sepsis. Among these fundamental issues, it is necessary to determine whether sepsis can present as a single entity or multiple entities. Additionally, what kind of sample should be used, and which biomarkers are the most reliable and valid for measuring adult/neonatal sepsis?
Immune dysregulation during bacterial sepsis
Sepsis can arise from an abnormal host immune response induced by an infection in any part of the body, particularly in the lungs and kidneys. There is no doubt that the interplay between the host and the virulence factors of the pathogen is a major cornerstone that determines the outcome of any infection. In this context, the balance between the pro-inflammatory and anti-inflammatory responses of the host is a vital determinant following pathogen invasion. The delicate balance between the competing pro-inflammatory and anti-inflammatory pathways determines the fate of patients with sepsis.[10,11] Over the last three decades, two conflicting theories have emerged regarding the host immune response during sepsis. The first theory proposes that the initial phase of sepsis is characterized by a hyper-inflammatory response followed by an extended immunosuppressive phase. On the other hand, the second more recent acceptable theory states that an anti-inflammatory response occurs simultaneously, counterbalancing the initial, more prominent hyper-inflammatory phase of sepsis.[11,12]
Upon exposure to invading pathogens, pathogen-associated molecular patterns (PAMPs), such as peptidoglycans, lipopolysaccharide (LPS), flagellin, bacterial DNA and the cell wall component of the gram-positive bacteria lipoteichoic acid, are recognized by pattern recognition proteins (PRPs), which are key components of the innate immune system. These PRPs can also recognize endogenous danger signals known as damage-associated molecular patterns (DAMPs), which are released during inflammatory stress. The interaction between PRP expressed by host cells and PAMPs expressed by pathogens leads to dimerization and conformational changes and triggers downstream signaling events via receptor/adaptor complexes such as myeloid differentiation factor 88 (MyD88) and Toll/interleukin 1 receptor domain-containing adapter-inducing interferon-β (TRIF). This process ultimately activates the nuclear factor-kappa B (NF-κB) transcription pathway, leading to a pro-inflammatory response, the magnitude of which depends on several factors, such as microbial load, drug resistance, virulence, age, host genetic polymorphisms, copy number variations, and comorbidities.[13⇓-15] This pro-inflammatory response is enhanced by complement (providing soluble PRP), leukocyte NETosis, endothelium and the coagulation system, and leads to the activation of the adaptive immune response.[6] In this stage, M1 macrophages excessively produce pro-inflammatory cytokines, which exacerbate immune damage. These cytokines include tumor necrosis factor (TNF)-α and interferon-γ (IFN-γ), as well as members of the interleukin (IL) family (IL-1, IL-3, IL-6, IL-8, etc.). The hyper-inflammatory response leads to the simultaneous activation of the anti-inflammatory response to restore immune homeostasis,[14] where M2 macrophages excessively produce high levels of anti-inflammatory cytokines, such as IL-4 and IL-10. In general, invading pathogens can be eliminated. However, if infection persists, the combined effects of pro- and anti-inflammatory responses that initially occur can harm cells and trigger the release of DAMPs, resulting in repeated cycles of continuous activation of a dysregulated immune response. As sepsis worsens, the inflammatory response progressively shifts from overactivation to immunosuppression, with decreasing immune cell numbers and immunological dysfunction being the predominant symptoms.[1,16,17] The increased expression of guanosine within the cerebrospinal fluid by pro-inflammatory cytokines increases the production of immature neutrophils into the bloodstream.[11] The natural killer (NK) cell count, cytokine output, and cytotoxic protein production from NK cells all decline.[18] Dendritic cell (DC) apoptosis, impaired maturation, and paralysis have been observed during this phase, contributing to reduction in the overall number of DCs. Additionally, alterations in surface molecules essential for DC function have also been reported, further promoting immune tolerance.[19] CD4+ T-cell counts decrease, and their functions, Th1/Th2 balance, and T-helper type 17 (Th17) / regulatory T (Treg) balance are disrupted.[1] Substintial alterations in B-cell counts and functions have also been reported.[20] As a result of the combined effects of these mechanisms, the body progressively loses its normal immune function and shifts into a state of immune paralysis or immunosuppression.[17]
Post-sepsis immunosuppression
Studies investigating the pathophysiological mechanisms of sepsis have shown that patients who die as a result of sepsis suffer from impaired host immunity.[21] The early stages of sepsis are predominated by a potent inflammatory response; hence, fever, shock and organ dysfunction occur in septic patients during this initial phase. Patients who survive the early stages of sepsis may develop nosocomial infections with organisms that are not typically pathogenic in immunocompetent hosts. The reason is that patients more often progress into a prolonged state of immune suppression in the later stages.[16] Postmortem examination of patients dying as a result of sepsis in ICUs revealed that the majority of these patients have unresolved septic foci, indicating that the invading microorganism was never eradicated.[22] Furthermore, postmortem studies revealed that post-sepsis immunosuppression occurs in multiple vital organs, such as lungs and spleen. Immunophenotyping of these two organs revealed the expansion of Treg cells and myeloid derived suppressor cells and the increased expression of several inhibitory receptors, such as programmed cell death 1 (PD-1). These findings indicate that sepsis results in the suppression of both innate and adaptive host immunity and thus leads to suppressed host immunity.[23] On the basis of these findings, augmentation of the host immune response is currently viewed as a promising therapeutic approach for sepsis.[24] Reprogramming of antigen-presenting cells and lymphocyte exhaustion are prominent features of sepsis-associated immune suppression. In addition to the depletion of T lymphocytes, postmortem studies have demonstrated that levels of IFN-γ and TNF produced by splenic T lymphocytes are lower than those produced by splenic T lymphocytes isolated from patients who die from other causes (non-infectious). Reprogramming of antigen-presenting cells involves reduced expression of human leukocyte antigen-DR (HLA-DR) and a decreased ability to release pro-inflammatory cytokines.[25]
Diagnostics of bacterial sepsis
Before sepsis treatment is initiated, blood cultures are conventionally taken, but this method has notable limitations, such as low sensitivity and delayed results.[26] Although the introduction of automated continuous blood culture monitoring systems has improved diagnostic reliability, subcultures remain necessary for further pathogen identification.[27] Treatment may then be adjusted.
Molecular microbiological assays, which detect the pathogen via amplification or hybridization methods, have been used as a suitable alternative to blood culture, especially in neonates where contamination, maternal antibiotic use and the challenge of obtaining sufficient blood samples can reduce the reliability of blood culture assays.[28] A systematic review and meta-analysis of 31 studies involving 5,920 patients with bacteraemia demonstrated that fast molecular diagnostic methods, such as peptide nucleic acid (PNA) fluorescence in situ hybridization (FISH), polymerase chain reaction (PCR), and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS), significantly lowered mortality risk when combined with an antimicrobial stewardship program.[29] In a study by Hayder Hamad et al,[30] PCR was evaluated as a diagnostic tool for detecting neonatal sepsis, with its effectiveness compared with that of blood culture in 85 neonates suspected of having sepsis. Interestingly, among the neonates who had a positive PCR result for 16S rRNA, 11 had negative results from bacterial blood cultures. Compared with traditional blood cultures, the use of a multiplex PCR test reduced the time required to obtain appropriate treatment in critically ill patients with culture-confirmed sepsis.[31] Although these molecular methods show promise, they may not provide sufficient diagnostic accuracy to fully replace microbial cultures.[32] The sensitivity of these assays is variable, and they face several technical and methodological challenges, including high cost and difficulties in integrating them into routine laboratory workflows. Additionally, a key limitation of these assays is that sepsis may be caused by a wide range of microorganisms, some of which are rare or sporadic, making it challenging to design a comprehensive assay that effectively detects all potential pathogens.[26] Manufacturers of these assays have adopted different strategies, using broadly conserved sequences (universal primers) or pathogen-specific assays (specific primers). However, both approaches can result in false-positive and false-negative results, respectively, which compromises the reliability of these assays and thereby stresses the need for new diagnostic approaches.[26,33]
In 2011, Pammi et al[34] conducted a systemic review and meta-analysis of 23 studies that addressed the use of microbiological molecular assays (PCR followed by direct sequencing) for the diagnosis of (neonatal) sepsis and concluded that these assays lack sufficient sensitivity to replace conventional blood culture methods. These molecular assays may be used in parallel (as “addon tests”) with microbiological cultures. The same group published a review of 35 studies that assessed the diagnostic accuracy of microbiological molecular assays in neonates with suspected sepsis. The molecular assays used in these studies were broad-range conventional PCR assays, realtime PCR, PCR followed by sequencing or hybridization, species-specific PCR assays, or multiplex PCR assays. There was substantial variation in the sensitivity of molecular assays in the diagnosis of neonatal sepsis among the reported studies. Patient characteristics and methods of DNA extraction are among the possible causes of variation in the sensitivity of these assays. Improvements in the diagnostic accuracy and development of molecular assays that evaluate antibiotic sensitivity are needed for these tests to play a substantial role in assisting the diagnosis and management of sepsis.[35]
Diagnostic profiling of patients with sepsis
Blood cultures, while traditionally used to diagnose bacterial sepsis, have notable limitations in reliability. This has led to a search for more predictive biomarkers as supplements to blood cultures that reflect the host’s immune response. Host-derived biomarkers, including gene expression profiles from whole blood samples, offer promising tools for understanding the causative pathogen, guiding treatment selection, and assessing prognosis. These biomarkers can also help categorize septic patients by mortality risk, enhancing patient stratification for more personalized care.[36,37]
However, meta-analysis caution against the causality of sepsis for long-term mortality, meaning that an outcome predictive remit of a biomarker for long-term survival is currently not achievable. Rather, age and comorbidities are of prognostic significance.[3]
Despite the availability of more than 178 sepsis-associated biomarkers, only a few of them are routinely used for the diagnosis of sepsis, including cytokine/chemokine biomarkers (IL-6, IL-8, IL-10, TNF-α), pro-inflammatory and acute-phase proteins (CRP, procalcitonin [PCT]), and coagulation profiles.[38,39]
For the diagnosis of sepsis, the use of marker panels rather than single markers is favored in transcriptomic and proteomic profiling studies.[40,41] Recent data have shown that a multi-marker approach can also help with prognosis prediction in patients with sepsis. PCT, presepsin, galectin-3, and soluble suppression of tumorigenicity 2 (sST2) are useful for predicting mortality in sepsis patients (analyzed in relation to 30-day all-cause mortality); thus, when combined with clinical variables, these parameters may be utilized to optimize the management of these patients.[42] In addition to PCT, nCD64 has been reported as a promising biomarker for the management of neonatal sepsis.[43] A study described microRNAs as “the answer to early diagnosis and staging of sepsis”. The unique microRNA expression pattern in sepsis was suggested as a tool for the diagnosis and management of sepsis. Nevertheless, more standardized and normalized data are warranted to confirm the diagnostic potential of microRNAs in sepsis.[44] The absence of timely diagnosis of sepsis has contributed to the increase in mortality among children in developing countries.[45,46]
Importantly, a wide spectrum of transcriptional changes occur in the leukocytes of septic patients. Genomic shifts usually occur within a few hours of the onset of sepsis, unlike other diseases, such as cancer, in which transcriptional shifts usually occur over an extended time frame. The rapid time frame of the genomic changes in sepsis complicates the process of comparing gene expression data of different septic patients,[47] as the exact stage of sepsis that has been sampled in each case is not known. Compared with healthy individuals, septic patients have 70%-80% of blood leukocytes RNA transcript significantly changed. These changes reflect the increased expression of the genes responsible for anti-inflammatory, pro-inflammatory and mitochondrial dysfunction pathways, in addition to the decreased expression of the genes responsible for translation initiation and antigen presentation. Interestingly, these alterations happen independently of the causative agent,[48] so they are unsuitable for the classification of bacterial sepsis (gram-positive or negative).
Selective adaptation of pathogens causing sepsis
Importantly, pathogens that cause sepsis (Table 1) undergo continuous adaptation after infecting their initial target tissues. This adaptation involves changes in the expression of virulence factors, allowing the pathogen to better evade the host’s immune response and persist within the host. The host relies on innate immune barriers as its first line of defense, which includes physical barriers, immune cells such as neutrophils and macrophages, and pattern recognition receptors. In the early stages of sepsis, immune activation predominantly involves the innate immune response. The adaptive immune response, which involves T and B cells, typically plays a more significant role in the later stages of infection. Both responses are influenced by other ongoing disease processes, treatments, and genetic factors, such as polymorphisms. However, pathogens often adapt faster, exploiting host factors to increase their survival. Genetic variants, including gene polymorphisms, single nucleotide polymorphisms (SNPs), haplogroups, and copy number variations, have been linked to susceptibility to sepsis.[49]
Table 1. Spectrum of bacteria causing sepsis in different patient groups
| Patient group | Bacteria | Reference |
|---|---|---|
| Neonates | ||
| Early onset (within the first 72 h) | Coagulase-negative staphylococci | [58] |
| Klebsiella pneumonia | [58, 59] | |
| Group B Streptococcus | [59, 60] | |
| Escherichia coli | [60] | |
| Listeria monocytogenes | [59] | |
| Streptococcus pneumoniae | [61] | |
| Late onset (after 72 h) | Coagulase-negative staphylococci, gram-negative bacilli, Enterobacteriaceae, and both methicillin- sensitive and methicillin-resistant Staphylococcus aureus | [62] |
| Children | Staphylococcus, Streptococcus, Pseudomonas, and Meningococcus | [63, 64] |
| Haemophilus influenza | [65] | |
| Proteus mirabilis | [65] | |
| Adults | Escherichia coli | [66] |
| Staphylococcus aureus | [67] | |
| Pseudomonas spp, Acinetobacter spp, Enterobacter spp | [68] | |
| Immunocompromised adults | Community acquired gram-negative bacteria, Staphylococcus aureus, gram-positive cocci (group A-β hemolytic Streptococcus that produces pyrogenic exotoxin) | [68] |
Pathogens are exposed to evolutionary selection pressures, producing a so-called bottleneck that ultimately affects the ability of a specific inoculum size to cause disease.[50] The sequential adaptations that occur in various environments —from the time point of infection to the manifestation of organ specific-disease and subsequent systemic spread — can lead to a reduction in genetic variation among bacterial populations, resulting in founder effects. For example, successive bottlenecks have been linked to the emergence of drug-resistant strains of Pseudomonas aeruginosa, one of the most problematic opportunistic human pathogens.[51] In contrast, Kono et al[52] reported no evidence of genetic adaptation when using a neonatal mouse model of colonization with a small inoculum of niche adapted S. pneumoniae (serotype 6). However, this study had limited time points for analysis and included viral co-infection. Various mechanisms contribute to the evolution of bacterial pathogens within a host, including mutations, phase variations, and horizontal gene transfers, which may be responsible for relapsing infections.[53]
Evidence of a single-clonal infective expansion
On the basis of a study carried out in mice, Gerlini et al[54] hypothesized that human pneumococcal bacteraemia generally arises from a single cell. This study used an intravenous infection model in which mice were inoculated with a mixture of three isogenic variants of S. pneumoniae. Genome sequencing revealed that in 4 out of the 6 cases, bacteraemia originated from a single bacterial cell. It has been suggested that the single cells initiating infection do not possess a virulence advantage over other cells in the challenge population. Furthermore, each cell in this population is thought to have an equal chance of establishing an infection. This study implies that among the cells that successfully withstand the host’s immune response, only one pneumococcus manages to establish bacteraemia. A shift toward increased heterogeneity has been observed following a single cell bottleneck, indicating positive selection for particular subclones. Another recent study examined how extraintestinal pathogenic Escherichia coli, a common cause of bacteremia, behaves during systemic infection in mice. While most organs are able to clear bacteria, some do not. This lack of clearance is mainly due to the significant growth of specific bacterial clones that initially made up less than 0.0001% of the original inoculum.[55] These findings indicate that although most bacterial clones are eliminated, certain clones can expand significantly in specific organs despite their initial low numbers. This situation demonstrates the bottleneck effect, where selective pressure reduces diversity, allowing only a few specific clones to survive and thrive. Pidwill et al[56] demonstrated that during Staphylococcus aureus infection, macrophages restrict bacterial growth, creating a bottleneck where only a small number of bacteria survive and replicate, leading to the formation of clonal abscesses that can spread to other parts of the body.
A study challenges the tight bottleneck theory in S. pneumoniae. By inoculating infant mice with an equal mixture of three isogenic S. pneumoniae mutants, Kono et al[52] reported that no single strain dominated the infection; instead, all three mutants were detected. There are notable differences between the two studies on S. pneumoniae, which offer different perspectives. The first study used adult mice, employed a high inoculum and included more time points, whereas the latter study utilized a neonatal mouse model with a lower inoculation dose and fewer time points. Notably, both studies utilized mouse models with laboratory-adapted strains grown to the logarithmic phase in defined broth, making it difficult to directly infer whether single clonal expansion occurs in humans.
Treatment of sepsis
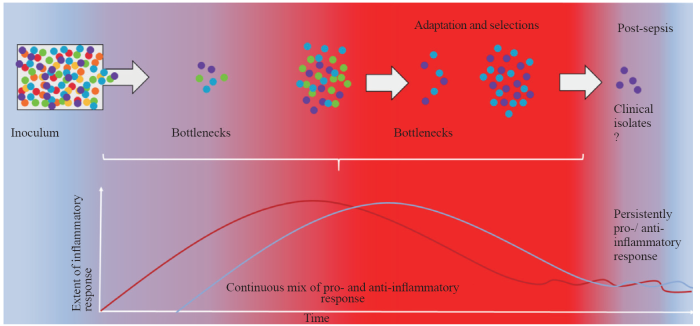
Early identification and management of sepsis within the first six hours of presentation are crucial for achieving optimal outcomes and minimizing associated mortality and morbidity. Initial management focuses on stabilizing respiration and providing fluid resuscitation, with continuous monitoring of vital signs, urine output and plasma lactate levels. This approach aims to restore circulation, ensure adequate oxygen delivery, and maintain tissue perfusion. Prompt identification and control of the source of infection, along with timely adjustment of treatment or surgical intervention, are crucial, as they significantly affect patient outcomes. It is strongly recommended that empiric anti-microbial therapy be initiated within the first hour, immediately after obtaining the microbiologic cultures (aerobic and anaerobic blood culture). Therapy should include one or more antimicrobials with activity against all likely pathogens and the ability to penetrate the tissues presumed to be the source of sepsis at adequate concentrations.[9] Multiple therapies are required, including glycemic control, deep venous thrombosis prophylaxis and measures to prevent anemia and coagulopathy, such as insulin, heparin and blood product therapy.[9,57] However, it remains challenging to assess how the timing of these treatments influences the dynamics and variability of the inflammatory response in a patient, which may coincide with the emergence of more virulent pathogenic strains (Figure 1). Some of these adapted strains can persist and contribute significantly to the long-term mortality of sepsis survivors.
Figure 1.
Figure 1.
Schematic presentation of parallel dynamics of selective pressures experienced by pathogens, which produce so-called bottlenecks, and the quality of the systemic inflammatory response. Recent experimental evidence has led to the development of a new sepsis model, challenging the traditional sepsis inflammatory concept in which an initial hyper-inflammatory response is followed by an immunosuppressive phase. This new sepsis model suggests the existence of a mixed inflammatory (heterogeneous) response in the initial phase of septicaemia.[69] The scale of the inflammatory response in sepsis can create a hostile environment for pathogens, applying strong selection pressure on the bacterial population (inoculum). This pressure may continuously result in a “bottleneck” effect, reducing the overall diversity of the bacterial population by allowing only specific clones with traits that enable them to survive immune assault to persist and proliferate. These surviving clones often possess adaptations, such as increased virulence factors or resistance to host defenses, which provide them with a survival advantage against immune system attack. Each pathogen is depicted as a colored sphere, with different colors representing distinct markers. The pro-inflammatory response is represented by the red line and red gradient, whereas the anti-inflammatory response is represented by the blue line and blue gradient.
CONCLUSION
Bacterial sepsis remains an important killer in all societies across all ages. While significant progress has been made in understanding the immunological mechanisms underlying the inflammatory response, there is growing recognition that the ongoing battle between host and pathogen, including the emergence of altered, adapted virulence, shapes both the acute and long-term response of patients with sepsis. This highlights the urgent need for novel high-throughput diagnostic methods and the development of more pre-emptive rather than reactive treatment strategies in sepsis care.
Funding: This research was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia, under grant number G-150-248-1443.
Ethical approval: Not needed.
Conflicts of interest: The authors declare that there is no conflict of interest.
Contributors: ASA proposed and wrote the paper. All authors contributed to the design and interpretation of the study and to further drafts.
Reference
Endothelial cell metabolism in sepsis
Sepsis heterogeneity
DOI:10.1007/s12519-023-00689-8
PMID:36735197
[Cited within: 1]

Sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection, with extremely high mortality. Notably, sepsis is a heterogeneous syndrome characterized by a vast, multidimensional array of clinical and biologic features, which has hindered advances in the therapeutic field beyond the current standards.We used PubMed to search the subject-related medical literature by searching for the following single and/or combination keywords: sepsis, heterogeneity, personalized treatment, host response, infection, epidemiology, mortality, incidence, age, children, sex, comorbidities, gene susceptibility, infection sites, bacteria, fungi, virus, host response, organ dysfunction and management.We found that host factors (age, biological sex, comorbidities, and genetics), infection etiology, host response dysregulation and multiple organ dysfunctions can all result in different disease manifestations, progression, and response to treatment, which make it difficult to effectively treat and manage sepsis patients.Herein, we have summarized contributing factors to sepsis heterogeneity, including host factors, infection etiology, host response dysregulation, and multiple organ dysfunctions, from the key elements of pathogenesis of sepsis. An in-depth understanding of the factors that contribute to the heterogeneity of sepsis will help clinicians understand the complexity of sepsis and enable researchers to conduct more personalized clinical studies for homogenous patients.© 2023. Children's Hospital, Zhejiang University School of Medicine.
Evidence for a causal link between sepsis and long-term mortality: a systematic review of epidemiologic studies
Diagnosis of bacterial sepsis: why are tests for bacteremia not sufficient
DOI:10.1080/14737159.2019.1660644 PMID:31446810 [Cited within: 1]
Estimates of sepsis prevalence and outcomes in adult patients in the ICU in India: a cross-sectional study
Immunopathophysiology of human sepsis
Update of sepsis in the intensive care unit
DOI:10.1159/000477419
PMID:28697503
[Cited within: 1]

Sepsis, the most common cause of admission to an intensive care unit (ICU), has had an increased incidence and prevalence over the last years with a simultaneous decrease in its short-term mortality. Sepsis survivors are more frequently discharged from hospital and often experience long-term outcomes such as late mortality, immune dysfunction, secondary infections, impaired quality of life, and unplanned readmissions. Early recognition and management of sepsis have challenged emergency care and critical care physicians and nurses. New sepsis definitions were produced and the Surviving Sepsis Campaign (SSC) 2016 was updated recently. Although hospital readmissions after sepsis are common, associated risk factors and how to manage patients who survive an episode of sepsis still need clarification. The immune dysfunction caused by sepsis/septic shock is complex, persistent, affects inflammatory and anti-inflammatory systems, and might be associated with long-term outcomes of sepsis. Several randomized controlled trials (RCT) that analyzed new (and old) interventions in sepsis/septic shock are discussed in this review in parallel with the SSC 2016 recommendations and other guidelines when relevant. RCTs addressing incidence, treatment, and prevention of important sepsis-associated organ dysfunction such as the acute respiratory distress syndrome, acute kidney injury, and brain dysfunction are highlighted. Finally, we briefly discuss the need for novel targets, predictive biomarkers, and new designs of RCTs in sepsis.© 2017 S. Karger AG, Basel.
Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet
DOI:S0140-6736(19)32989-7
PMID:31954465
[Cited within: 1]

Sepsis is life-threatening organ dysfunction due to a dysregulated host response to infection. It is considered a major cause of health loss, but data for the global burden of sepsis are limited. As a syndrome caused by underlying infection, sepsis is not part of standard Global Burden of Diseases, Injuries, and Risk Factors Study (GBD) estimates. Accurate estimates are important to inform and monitor health policy interventions, allocation of resources, and clinical treatment initiatives. We estimated the global, regional, and national incidence of sepsis and mortality from this disorder using data from GBD 2017.We used multiple cause-of-death data from 109 million individual death records to calculate mortality related to sepsis among each of the 282 underlying causes of death in GBD 2017. The percentage of sepsis-related deaths by underlying GBD cause in each location worldwide was modelled using mixed-effects linear regression. Sepsis-related mortality for each age group, sex, location, GBD cause, and year (1990-2017) was estimated by applying modelled cause-specific fractions to GBD 2017 cause-of-death estimates. We used data for 8·7 million individual hospital records to calculate in-hospital sepsis-associated case-fatality, stratified by underlying GBD cause. In-hospital sepsis-associated case-fatality was modelled for each location using linear regression, and sepsis incidence was estimated by applying modelled case-fatality to sepsis-related mortality estimates.In 2017, an estimated 48·9 million (95% uncertainty interval [UI] 38·9-62·9) incident cases of sepsis were recorded worldwide and 11·0 million (10·1-12·0) sepsis-related deaths were reported, representing 19·7% (18·2-21·4) of all global deaths. Age-standardised sepsis incidence fell by 37·0% (95% UI 11·8-54·5) and mortality decreased by 52·8% (47·7-57·5) from 1990 to 2017. Sepsis incidence and mortality varied substantially across regions, with the highest burden in sub-Saharan Africa, Oceania, south Asia, east Asia, and southeast Asia.Despite declining age-standardised incidence and mortality, sepsis remains a major cause of health loss worldwide and has an especially high health-related burden in sub-Saharan Africa.The Bill & Melinda Gates Foundation, the National Institutes of Health, the University of Pittsburgh, the British Columbia Children's Hospital Foundation, the Wellcome Trust, and the Fleming Fund.Copyright © 2020 The Author(s). Published by Elsevier Ltd. This is an Open Access Article under the CC BY 4.0 licence. Published by Elsevier Ltd.. All rights reserved.
Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021
DOI:10.1007/s00134-021-06506-y PMID:34599691 [Cited within: 3]
Sepsis: inflammation is a necessary evil
DOI:10.3389/fcell.2019.00108
PMID:31281814
[Cited within: 1]

Sepsis is one of the leading causes of deaths world-wide and yet there are no therapies available other than ICU treatment. The patient outcome is determined by a complex interplay between the pro and anti-inflammatory responses of the body i.e., a homeostatic balance between these two competing events to be achieved for the patient's recovery. The initial attempts on drug development mainly focused on controlling inflammation, however, without any tangible outcome. This was despite most deaths occurring during the immune paralysis stage of this biphasic disease. Recently, the focus has been shifting to understand immune paralysis (caused by apoptosis and by anti-inflammatory cytokines) to develop therapeutic drugs. In this review we put forth an argument for a proper understanding of the molecular basis of inflammation as well as apoptosis for developing an effective therapy.
Neutrophil dysregulation during sepsis: an overview and update
Sepsis: current definition, pathophysiology, diagnosis, and management
DOI:10.1177/0884533617695243
PMID:28537517
[Cited within: 1]

Sepsis is a clinical syndrome that results from the dysregulated inflammatory response to infection that leads to organ dysfunction. The resulting losses to society in terms of financial burden, morbidity, and mortality are enormous. We provide a review of sepsis, its underlying pathophysiology, and guidance for diagnosis and management of this common disease. Current established treatments include appropriate antimicrobial agents to target the underlying infection, optimization of intravascular volume to improve stroke volume, vasopressors to counteract vasoplegic shock, and high-quality supportive care. Appropriate implementation of established treatments combined with novel therapeutic approaches promises to continue to decrease the impact of this disease.
Microbial recognition and danger signals in sepsis and trauma
Immune response in bacterial and Candida sepsis
Evolving paradigms in sepsis management: a narrative review
Role of the adaptive immune response in sepsis
DOI:10.1186/s40635-020-00309-z
PMID:33336293
[Cited within: 2]

Sepsis is a syndrome of shock and dysfunction of multiple vital organs that is caused by an uncontrolled immune response to infection and has a high mortality rate. There are no therapies for sepsis, and it has become a global cause for concern. Advances in patient care and management now mean that most patients survive the initial hyper-inflammatory phase of sepsis but progress to a later immunosuppressed phase, where 30% of patients die due to secondary infection. Deficits in the adaptive immune response may play a major role in sepsis patient mortality. The adaptive immune response involves a number of cell types including T cells, B cells and dendritic cells, all with immunoregulatory roles aimed at limiting damage and returning immune homeostasis after infection or insult. However, in sepsis, adaptive immune cells experience cell death or exhaustion, meaning that they have defective effector and memory responses ultimately resulting in an ineffective or suppressed immune defence. CD4+ T cells seem to be the most susceptible to cell death during sepsis and have ensuing defective secretory profiles and functions. Regulatory T cells seem to evade apoptosis and contribute to the immune suppression observed with sepsis. Preclinical studies have identified a number of new targets for therapy in sepsis including anti-apoptotic agents and monoclonal antibodies aimed at reducing cell death, exhaustion and maintaining/restoring adaptive immune cell functions. While early phase clinical trials have demonstrated safety and encouraging signals for biologic effect, larger scale clinical trial testing is required to determine whether these strategies will prove effective in improving outcomes from sepsis.
The biology of natural killer cells during sepsis
DOI:10.1111/imm.12854
PMID:29064085
[Cited within: 1]

Natural killer (NK) cells are large granular lymphocytes largely recognized for their importance in tumour surveillance and the host response to viral infections. However, as the major innate lymphocyte population, NK cells also coordinate early responses to bacterial infections by amplifying the antimicrobial functions of myeloid cells, especially macrophages, by production of interferon-γ (IFN-γ). Alternatively, excessive NK cell activation and IFN-γ production can amplify the systemic inflammatory response during sepsis resulting in increased physiological dysfunction and organ injury. Our understanding of NK cell biology during bacterial infections and sepsis is mostly derived from studies performed in mice. Human studies have demonstrated a correlation between altered NK cell functions and outcomes during sepsis. However, mechanistic understanding of NK cell function during human sepsis is limited. In this review, we will review the current understanding of NK cell biology during sepsis and discuss the challenges associated with modulating NK cell function during sepsis for therapeutic benefit.© 2017 John Wiley & Sons Ltd.
Dendritic cells in sepsis: Potential immunoregulatory cells with therapeutic potential
DOI:S0161-5890(18)30534-0
PMID:30007546
[Cited within: 1]

Sepsis is a disease of dysfunctional immune response against the pathogen causing a profound immune-mediated damage to the vital organs and death of the patient in most cases. However, when sepsis is described much attention is given to monocytes/macrophages, complement system, neutrophils, cytokine storm, and T cells. Dendritic cells (DCs) get less attention in this scenario despite comprising the major immune cell population. Therefore the present review is designed to highlight the importance of DCs in the pathogenesis of sepsis, sepsis-associated immunosuppression, and organ damage. The article starts with an introduction of sepsis as a major medical problem needing an urgent therapeutic targeting. Thereafter it provides a brief information regarding classical and plasmacytoid DCs and their role in the maintenance of immune homeostasis. The subsequent sections describe the role of DCs in the immunopathogenesis of sepsis via immunoregulation, impact of sepsis on DCs including their immunometabolic changes, and their therapeutic targeting during sepsis.Copyright © 2018 Elsevier Ltd. All rights reserved.
The emerging roles and therapeutic potential of B cells in sepsis
The immune system's role in sepsis progression, resolution, and long-term outcome
DOI:10.1111/imr.12499
PMID:27782333
[Cited within: 1]

Sepsis occurs when an infection exceeds local tissue containment and induces a series of dysregulated physiologic responses that result in organ dysfunction. A subset of patients with sepsis progress to septic shock, defined by profound circulatory, cellular, and metabolic abnormalities, and associated with a greater mortality. Historically, sepsis-induced organ dysfunction and lethality were attributed to the complex interplay between the initial inflammatory and later anti-inflammatory responses. With advances in intensive care medicine and goal-directed interventions, early 30-day sepsis mortality has diminished, only to steadily escalate long after "recovery" from acute events. As so many sepsis survivors succumb later to persistent, recurrent, nosocomial, and secondary infections, many investigators have turned their attention to the long-term sepsis-induced alterations in cellular immune function. Sepsis clearly alters the innate and adaptive immune responses for sustained periods of time after clinical recovery, with immune suppression, chronic inflammation, and persistence of bacterial representing such alterations. Understanding that sepsis-associated immune cell defects correlate with long-term mortality, more investigations have centered on the potential for immune modulatory therapy to improve long-term patient outcomes. These efforts are focused on more clearly defining and effectively reversing the persistent immune cell dysfunction associated with long-term sepsis mortality.© 2016 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Sepsis-induced immune alterations monitoring by flow cytometry as a promising tool for individualized therapy
Immunosuppression in patients who die of sepsis and multiple organ failure
DOI:10.1001/jama.2011.1829
PMID:22187279
[Cited within: 1]

Severe sepsis is typically characterized by initial cytokine-mediated hyperinflammation. Whether this hyperinflammatory phase is followed by immunosuppression is controversial. Animal studies suggest that multiple immune defects occur in sepsis, but data from humans remain conflicting.To determine the association of sepsis with changes in host innate and adaptive immunity and to examine potential mechanisms for putative immunosuppression.Rapid postmortem spleen and lung tissue harvest was performed at the bedsides of 40 patients who died in intensive care units (ICUs) of academic medical centers with active severe sepsis to characterize their immune status at the time of death (2009-2011). Control spleens (n = 29) were obtained from patients who were declared brain-dead or had emergent splenectomy due to trauma; control lungs (n = 20) were obtained from transplant donors or from lung cancer resections.Cytokine secretion assays and immunophenotyping of cell surface receptor-ligand expression profiles were performed to identify potential mechanisms of immune dysfunction. Immunohistochemical staining was performed to evaluate the loss of immune effector cells.The mean ages of patients with sepsis and controls were 71.7 (SD, 15.9) and 52.7 (SD, 15.0) years, respectively. The median number of ICU days for patients with sepsis was 8 (range, 1-195 days), while control patients were in ICUs for 4 or fewer days. The median duration of sepsis was 4 days (range, 1-40 days). Compared with controls, anti-CD3/anti-CD28-stimulated splenocytes from sepsis patients had significant reductions in cytokine secretion at 5 hours: tumor necrosis factor, 5361 (95% CI, 3327-7485) pg/mL vs 418 (95% CI, 98-738) pg/mL; interferon γ, 1374 (95% CI, 550-2197) pg/mL vs 37.5 (95% CI, -5 to 80) pg/mL; interleukin 6, 3691 (95% CI, 2313-5070) vs 365 (95% CI, 87-642) pg/mL; and interleukin 10, 633 (95% CI, -269 to 1534) vs 58 (95% CI, -39 to 156) pg/mL; (P <.001 for all). There were similar reductions in 5-hour lipopolysaccharide-stimulated cytokine secretion. Cytokine secretion in sepsis patients was generally less than 10% that in controls, independent of age, duration of sepsis, corticosteroid use, and nutritional status. Although differences existed between spleen and lung, flow cytometric analysis showed increased expression of selected inhibitory receptors and ligands and expansion of suppressor cell populations in both organs. Unique differences in cellular inhibitory molecule expression existed in immune cells isolated from lungs of sepsis patients vs cancer patients and vs transplant donors. Immunohistochemical staining showed extensive depletion of splenic CD4, CD8, and HLA-DR cells and expression of ligands for inhibitory receptors on lung epithelial cells.Patients who die in the ICU following sepsis compared with patients who die of nonsepsis etiologies have biochemical, flow cytometric, and immunohistochemical findings consistent with immunosuppression. Targeted immune-enhancing therapy may be a valid approach in selected patients with sepsis.
Sepsis-induced immunosuppression
The immunopathology of sepsis and potential therapeutic targets
DOI:10.1038/nri.2017.36
PMID:28436424
[Cited within: 1]

Sepsis is defined as a life-threatening organ dysfunction that is caused by a dysregulated host response to infection. In sepsis, the immune response that is initiated by an invading pathogen fails to return to homeostasis, thus culminating in a pathological syndrome that is characterized by sustained excessive inflammation and immune suppression. Our understanding of the key mechanisms involved in the pathogenesis of sepsis has increased tremendously, yet this still needs to be translated into novel targeted therapeutic strategies. Pivotal for the clinical development of new sepsis therapies is the selection of patients on the basis of biomarkers and/or functional defects that provide specific insights into the expression or activity of the therapeutic target.
Emerging technologies for molecular diagnosis of sepsis
Development of new methods for detecting bloodstream pathogens
Diagnostics for neonatal sepsis: current approaches and future directions
Role of rapid diagnostics in diagnosis and management of patients with sepsis
Comparison between polymerase chain reaction and blood culture for diagnosis of neonatal sepsis
DOI:10.22092/ARI.2022.358608.2259
PMID:37312696
[Cited within: 1]

Neonatal sepsis can be defined as any systemic bacterial infection confirmed by a positive blood culture in the first month of life. This study evaluated the polymerase chain reaction as the diagnostic approach to identify neonatal sepsis instead of blood culture. In this study, 85 blood specimens were collected from 85 patients with suspected septicemia; ages ranged between 1 to 28 days from both sexes (53 males and 32 females) from November 2014 to March 2015. From each neonate, a minimum of 1-3 ml of blood was collected by standard sterile procedures, 2 ml for blood culture, while 1 ml was used for DNA extraction. A minimum of 2 ml of blood is taken through venipuncture and injected into two or more "blood bottles" with specific media for aerobic and anaerobic organisms. The blood is collected using an aseptic technique. The recorded data showed that the bacterial culture was positive in 7.06% of patients versus 92.9%, revealing a negative bacterial culture. The most common types of bacteria isolated were three isolates of spp. (50.0%), followed by one isolate of (16.67%), one (16.67%) isolate, and one spp. (16.67%) isolate. Finally, molecular detection for bacterial sepsis was done using specific primers (, and ). It was found that genes were present in 20% of samples, and gene was present in (18.8%). While its gene used for the detection of fungi revealed negative results in all samples.
Blood gas analysis as a surrogate for microhemodynamic monitoring in sepsis
DOI:10.5847/wjem.j.1920-8642.2023.093
PMID:37969221
[Cited within: 1]

Emergency patients with sepsis or septic shock are at high risk of death. Despite increasing attention to microhemodynamics, the clinical use of advanced microcirculatory assessment is limited due to its shortcomings. Since blood gas analysis is a widely used technique reflecting global oxygen supply and consumption, it may serve as a surrogate for microcirculation monitoring in septic treatment.We performed a search using PubMed, Web of Science, and Google scholar. The studies and reviews that were most relevant to septic microcirculatory dysfunctions and blood gas parameters were identified and included.Based on the pathophysiology of oxygen metabolism, the included articles provided a general overview of employing blood gas analysis and its derived set of indicators for microhemodynamic monitoring in septic care. Notwithstanding flaws, several parameters are linked to changes in the microcirculation. A comprehensive interpretation of blood gas parameters can be used in order to achieve hemodynamic optimization in septic patients.Blood gas analysis in combination with clinical performance is a reliable alternative for microcirculatory assessments. A deep understanding of oxygen metabolism in septic settings may help emergency physicians to better use blood gas analysis in the evaluation and treatment of sepsis and septic shock.Copyright: © World Journal of Emergency Medicine.
Application of advanced molecular methods to study early-onset neonatal sepsis
The challenge of molecular diagnosis of bloodstream infections
Molecular assays in the diagnosis of neonatal sepsis: a systematic review and meta-analysis
Molecular assays for the diagnosis of sepsis in neonates
Biomarkers of inflammation and the etiology of sepsis
Diagnosing and managing sepsis by probing the host response to infection: advances, opportunities, and challenges
Biomarkers for sepsis: more than just fever and leukocytosis-a narrative review
Biomarkers of sepsis: time for a reappraisal
Combined transcriptome and proteome leukocyte’s profiling reveals up-regulated module of genes/proteins related to low density neutrophils and impaired transcription and translation processes in clinical sepsis
Leveraging transcriptomics for precision diagnosis: lessons learned from cancer and sepsis
Multi-marker approach using procalcitonin, presepsin, galectin-3, and soluble suppression of tumorigenicity 2 for the prediction of mortality in sepsis
DOI:10.1186/s13613-017-0252-y
PMID:28271449
[Cited within: 1]

Background: Biomarker could be objective and reliable tools to predict mortality in sepsis. We explored the prognostic utilities of emerging biomarkers in septic patients and questioned whether adding biomarkers to the clinical variables would improve the prediction of mortality in sepsis.Methods: This retrospective study included 157 septic patients (112 patients with sepsis; 45 patients with septic shock). Procalcitonin (PCT), presepsin, galectin-3, and soluble suppression of tumorigenicity 2 (sST2) concentrations were analyzed in relation to the 30-day all-cause mortality. Their value added on top of Sequential (Sepsis-related) Organ Failure Assessment (SOFA) score, high-sensitivity C-reactive protein, and white blood cells was also analyzed.Results: PCT could not predict 30-day mortality. Univariate hazard ratio [HR with 95% confidence interval (CI)] of the other dichotomized variables was: 1.33 (0.55-3.194) for presepsin; 7.87 (2.29-26.96) for galectin-3; 1.55 (0.71-3.38) for sST2; and 2.18 (1.01-4.75) for SOFA score. The risk of 30-day mortality increased stepwise as the number of biomarkers above optimal cutoff values increased, and the highest risk was observed when all four biomarkers and SOFA score increased (HR = 14.5). Multi-marker approach predicted 30-day mortality better than SOFA score [area under the curves (95% CI), 0.769 (0.695-0.833) vs. 0.615 (0.535-0.692)]. In reclassification analyses, adding biomarkers to clinical variables improved the prediction of mortality.Conclusion: This study demonstrated a possible prognostic utility of PCT, presepsin, galectin-3, and sST2 in sepsis. Multi-marker approach could be beneficial for an optimized management of patients with sepsis.
Biomarkers for the diagnosis of neonatal sepsis and necrotizing enterocolitis: clinical practice guidelines
DOI:S0378-3782(16)30564-3
PMID:28131458
[Cited within: 1]

Sepsis and necrotizing enterocolitis are major contributors to morbidity and mortality in neonates, especially in those born preterm. While therapeutic interventions are available for both (for e.g. antibiotics), a major dilemma is early diagnosis so that these interventions can be done in a timely manner. As clinical evaluation alone is unreliable in identifying infants in the early stages of neonatal sepsis or necrotizing enterocolitis, there is a need to find specific biomarkers associated with these conditions to improve diagnostic capabilities. Optimal use of biomarkers in the identification and management of affected neonates requires an understanding of the properties of each marker within the timeline of the inflammatory response. We propose that early- and mid-phase markers such as neutrophil CD64 and procalcitonin should be combined with the late-phase biomarker C-reactive protein for maximal diagnostic benefit. Appropriately powered trials evaluating the serial measurements of these markers in decisions related to antibiotic stewardship in the neonatal population are indicated, in addition to more studies investigating other potentially useful biomarkers.Copyright © 2016. Published by Elsevier B.V.
Role of microRNAs in sepsis
DOI:10.1007/s00011-017-1031-9
PMID:28258291
[Cited within: 1]

MicroRNAs have been found to be of high significance in the regulation of various genes and processes in the body. Sepsis is a serious clinical problem which arises due to the excessive host inflammatory response to infection. The non-specific clinical features and delayed diagnosis of sepsis has been a matter of concern for long time.MicroRNAs could enable better diagnosis of sepsis and help in the identification of the various stages of sepsis. Improved diagnosis may enable quicker and more effective treatment measures. The initial acute and transient phase of sepsis involves excessive secretion of pro-inflammatory cytokines which causes severe damage. MicroRNAs negatively regulate the toll-like receptor signaling pathway and regulate the production of inflammatory cytokines during sepsis. Likewise, microRNAs have shown to regulate the vascular barrier and endothelial function in sepsis. They are also involved in the regulation of the apoptosis, immunosuppression, and organ dysfunction in later stages of sepsis. Their importance at various levels of the pathophysiology of sepsis has been discussed along with the challenges and future perspectives.MicroRNAs could be key players in the diagnosis and staging of sepsis. Their regulation at various stages of sepsis suggests that they may have an important role in altering the outcome associated with sepsis.
Identifying patients with sepsis on the hospital wards
DOI:S0012-3692(16)50385-7
PMID:27374948
[Cited within: 1]

Sepsis contributes to up to half of all deaths in hospitalized patients, and early interventions, such as appropriate antibiotics, have been shown to improve outcomes. Most research has focused on early identification and treatment of patients with sepsis in the ED and the ICU; however, many patients acquire sepsis on the general wards. The goal of this review is to discuss recent advances in the detection of sepsis in patients on the hospital wards. We discuss data highlighting the benefits and limitations of the systemic inflammatory response syndrome (SIRS) criteria for screening patients with sepsis, such as its low specificity, as well as newly described scoring systems, including the proposed role of the quick sepsis-related organ failure assessment (qSOFA) score. Challenges specific to detecting sepsis on the wards are discussed, and future directions that use big data approaches and automated alert systems are highlighted.Copyright © 2016 American College of Chest Physicians. Published by Elsevier Inc. All rights reserved.
The burden of sepsis-a call to action in support of World Sepsis Day 2013
DOI:10.1016/j.jcrc.2013.04.012 PMID:23747158 [Cited within: 1]
Gene expression profiling in sepsis: timing, tissue, and translational considerations
DOI:10.1016/j.molmed.2014.01.006
PMID:24548661
[Cited within: 1]

Sepsis is a complex inflammatory response to infection. Microarray-based gene expression studies of sepsis have illuminated the complex pathogen recognition and inflammatory signaling pathways that characterize sepsis. More recently, gene expression profiling has been used to identify diagnostic and prognostic gene signatures, as well as novel therapeutic targets. Studies in pediatric cohorts suggest that transcriptionally distinct subclasses might account for some of the heterogeneity seen in sepsis. Time series analyses have pointed to rapid and dynamic shifts in transcription patterns associated with various phases of sepsis. These findings highlight current challenges in sepsis knowledge translation, including the need to adapt complex and time-consuming whole-genome methods for use in the intensive care unit environment, where rapid diagnosis and treatment are essential. Copyright © 2014 Elsevier Ltd. All rights reserved.
Shared and distinct aspects of the sepsis transcriptomic response to fecal peritonitis and pneumonia
Sepsis genomics: stepping forward toward sepsis prevention
Analysis of bottlenecks in experimental models of infection
Bottleneck size and selection level reproducibly impact evolution of antibiotic resistance
DOI:10.1038/s41559-021-01511-2
PMID:34312522
[Cited within: 1]

During antibiotic treatment, the evolution of bacterial pathogens is fundamentally affected by bottlenecks and varying selection levels imposed by the drugs. Bottlenecks-that is, reductions in bacterial population size-lead to an increased influence of random effects (genetic drift) during bacterial evolution, and varying antibiotic concentrations during treatment may favour distinct resistance variants. Both aspects influence the process of bacterial evolution during antibiotic therapy and thereby treatment outcome. Surprisingly, the joint influence of these interconnected factors on the evolution of antibiotic resistance remains largely unexplored. Here we combine evolution experiments with genomic and genetic analyses to demonstrate that bottleneck size and antibiotic-induced selection reproducibly impact the evolutionary path to resistance in pathogenic Pseudomonas aeruginosa, one of the most problematic opportunistic human pathogens. Resistance is favoured-expectedly-under high antibiotic selection and weak bottlenecks, but-unexpectedly-also under low antibiotic selection and severe bottlenecks. The latter is likely to result from a reduced probability of losing favourable variants through drift under weak selection. Moreover, the absence of high resistance under low selection and weak bottlenecks is caused by the spread of low-resistance variants with high competitive fitness under these conditions. We conclude that bottlenecks, in combination with drug-induced selection, are currently neglected key determinants of pathogen evolution and outcome of antibiotic treatment.© 2021. The Author(s).
Single cell bottlenecks in the pathogenesis of Streptococcus pneumoniae
Within-host evolution of bacterial pathogens
DOI:10.1038/nrmicro.2015.13
PMID:26806595
[Cited within: 1]

Whole-genome sequencing has opened the way for investigating the dynamics and genomic evolution of bacterial pathogens during the colonization and infection of humans. The application of this technology to the longitudinal study of adaptation in an infected host--in particular, the evolution of drug resistance and host adaptation in patients who are chronically infected with opportunistic pathogens--has revealed remarkable patterns of convergent evolution, suggestive of an inherent repeatability of evolution. In this Review, we describe how these studies have advanced our understanding of the mechanisms and principles of within-host genome evolution, and we consider the consequences of findings such as a potent adaptive potential for pathogenicity. Finally, we discuss the possibility that genomics may be used in the future to predict the clinical progression of bacterial infections and to suggest the best option for treatment.
The role of host and microbial factors in the pathogenesis of pneumococcal bacteraemia arising from a single bacterial cell bottleneck
Pathogen clonal expansion underlies multiorgan dissemination and organ-specific outcomes during murine systemic infection
Clonal population expansion of Staphylococcus aureus occurs due to escape from a finite number of intraphagocyte niches
DOI:10.1038/s41598-023-27928-2
PMID:36681703
[Cited within: 1]

Staphylococcus aureus is a human commensal and also an opportunist pathogen causing life threatening infections. During S. aureus disease, the abscesses that characterise infection can be clonal, whereby a large bacterial population is founded by a single or few organisms. Our previous work has shown that macrophages are responsible for restricting bacterial growth such that a population bottleneck occurs and clonality can emerge. A subset of phagocytes fail to control S. aureus resulting in bacterial division, escape and founding of microabscesses that can seed other host niches. Here we investigate the basis for clonal microabscess formation, using in vitro and in silico models of S. aureus macrophage infection. Macrophages that fail to control S. aureus are characterised by formation of intracellular bacterial masses, followed by cell lysis. High-resolution microscopy reveals that most macrophages had internalised only a single S. aureus, providing a conceptual framework for clonal microabscess generation, which was supported by a stochastic individual-based, mathematical model. Once a threshold of masses was reached, increasing the number of infecting bacteria did not result in greater mass numbers, despite enhanced phagocytosis. This suggests a finite number of permissive, phagocyte niches determined by macrophage associated factors. Increased understanding of the parameters of infection dynamics provides avenues for development of rational control measures.© 2023. The Author(s).
Distribution, antimicrobial resistance and predictors of mortality in neonatal sepsis
DOI:10.3233/NPM-1765
PMID:29991144
[Cited within: 2]

The aim of this study is to investigate etiological agents, patterns of antimicrobial resistance and predictors of mortality in culture proven neonatal sepsis.This is a twenty-four month retrospective cohort study of infants with culture proven sepsis. Demographic data, type of isolates and its sensitivity pattern were recorded. Multidrug resistant gram-negative isolates were defined as resistance to any three of five antibiotic classes: extended-spectrum cephalosporins, carbapenems, aminoglycosides, fluoroquinolones and piperacillin-tazobactam.A total of 183 case with culture positive sepsis were identified. Early onset sepsis occurred in 59% of cases. The majority of isolates (56.2%) were gram-positive but the most common individual isolates were klebsiella spp. (31.1%), Staphylococcus aureus (24.5%) and coagulase-negative staphylococci (CONS) (22.9%). The pathogen mix in early-onset did not differ from late-onset sepsis. High rates of multidrug resistance were observed in klebsiella spp. (49.1%), Escherichia coli (50%), citrobacter spp (50%), acinetobacter spp. (28.5%), pseudomonas spp. (100%) isolates. Methicillin resistance prevailed in 16.6% of coagulase-negative staphylococci, 24.4% of Staphylococcus aureus and 62.5% of enterococcus spp. Multivariate analysis revealed invasive ventilation and early onset sepsis to be independently associated with increased risk of mortality in contrast to breast milk feeding which is associated with decreased risk of mortality.A high degree of antimicrobial resistance underscores the need to understand the pathogenesis of resistance, curtail the irrational prescription of antibiotics in neonates and the requirement for measures to prevent it in low-income and middle-income countries.
Pediatric sepsis update: how are children different
Pediatric sepsis
Early-onset neonatal sepsis caused by Streptococcus pneumoniae serogroup 8
Br J Hosp Med (Lond).
Neonatal sepsis
DOI:S0140-6736(17)31002-4
PMID:28434651
[Cited within: 1]

Neonatal sepsis is the cause of substantial morbidity and mortality. Precise estimates of neonatal sepsis burden vary by setting. Differing estimates of disease burden have been reported from high-income countries compared with reports from low-income and middle-income countries. The clinical manifestations range from subclinical infection to severe manifestations of focal or systemic disease. The source of the pathogen might be attributed to an in-utero infection, acquisition from maternal flora, or postnatal acquisition from the hospital or community. The timing of exposure, inoculum size, immune status of the infant, and virulence of the causative agent influence the clinical expression of neonatal sepsis. Immunological immaturity of the neonate might result in an impaired response to infectious agents. This is especially evident in premature infants whose prolonged stays in hospital and need for invasive procedures place them at increased risk for hospital-acquired infections. Clinically, there is often little difference between sepsis that is caused by an identified pathogen and sepsis that is caused by an unknown pathogen. Culture-independent diagnostics, the use of sepsis prediction scores, judicious antimicrobial use, and the development of preventive measures including maternal vaccines are ongoing efforts designed to reduce the burden of neonatal sepsis.Copyright © 2017 Elsevier Ltd. All rights reserved.
Epidemiology of blood culture-proven bacterial sepsis in children in Switzerland: a population-based cohort study
DOI:S2352-4642(17)30010-X
PMID:30169202
[Cited within: 1]

Sepsis is a leading cause of childhood mortality worldwide. We assessed population-based incidence and outcomes of blood culture-proven bacterial sepsis in children in Switzerland.We did a multicentre, prospective, cohort study at ten paediatric hospitals in Switzerland. We included neonates and children younger than 17 years with blood culture-proven bacterial sepsis. Children were eligible if they met criteria for systemic inflammatory response syndrome-according to 2005 paediatric consensus definition- at the time of blood culture sampling. Incidence was calculated by dividing the number of annual sepsis episodes in the study for the years 2012-15 by the end-of-year resident paediatric population in Switzerland. The primary outcome was in-hospital mortality in the first 30 days after sepsis onset.Between Sept 1, 2011, and Dec 31, 2015, we enrolled 1096 children to our study. Of 1181 episodes of blood culture-proven bacterial sepsis, 382 (32%) occurred in 379 previously healthy children, 402 (34%) in 391 neonates, and 397 (34%) in 341 children with comorbidities. Incidence was 25·1 cases per 100 000 (95% CI 23·8-26·4) in children and 146·0 cases per 100 000 (133·2-159·6) in neonates. Central line-associated bloodstream infections and primary bloodstream infections accounted for 569 (48%) of 1181 episodes, and organ dysfunction was present in 455 (39%) of 1181 episodes. Escherichia coli (242 of 1181 [20%]), Staphylococcus aureus (177 of 1181 [15%]), coagulase-negative staphylococci (135 of 1181 [11%]), and Streptococcus pneumoniae (118 of 1181 [10%]) were the most prevalent pathogens in our study, accounting for 57% of episodes. The overall case-fatality ratio was 7% (82 of 1181 episodes; 95% CI 5·6-8·6), and it was higher in neonates (11%, 45 of 402 episodes; 8·4-14·8; adjusted odds ratio [OR] 4·41, 95% CI 1·75-11·1) and children with comorbidities (7%, 27 of 397 episodes; 4·6-9·9; OR 4·97, 1·84-13·4) compared with previously healthy children (3%, ten of 382 episodes; 1·3-4·9). The case-fatality ratio was 1% (five of 726 episodes [95% CI 0·3-1·7]) for children without organ dysfunction, which increased to 17% (77 of 455 episodes [13·7-20·8]) when organ dysfunction was present (adjusted OR 4·84, 95% CI 1·40-16·7).The burden of blood culture-proven bacterial sepsis on child health remains considerable. We recorded key differences in predominant organisms, severity, and outcome between neonates, previously healthy children, and children with comorbidities. Although for most episodes of blood culture-proven bacterial sepsis, no organ dysfunction was seen, presence of organ dysfunction was strongly associated with mortality.Swiss National Science Foundation, Swiss Society of Intensive Care, Bangerter Foundation, Vinetum and Borer Foundation, and Foundation for the Health of Children and Adolescents.Copyright © 2017 Elsevier Ltd. All rights reserved.
Bacterial and fungal etiology of sepsis in children in the United States: reconsidering empiric therapy
Pediatric severe sepsis: current trends and outcomes from the Pediatric Health Information Systems database
Pediatr Crit Care Med.
Outcomes of older adults with sepsis at admission to an intensive care unit
Bacterial and fungal profile, drug resistance pattern and associated factors of isolates recovered from blood samples of patients referred to Ethiopian Public Health Institute: cross-sectional study
DOI:10.1186/s12879-021-06896-w
PMID:34844570
[Cited within: 1]

Blood stream infections are serious infections that usually induce prolongation of hospital stay, morbidity and mortality in several countries including Ethiopia. The aim of this study was to determine bacterial and fungal profile, their drug resistance patterns, and risk factors associated with blood stream infections.A cross sectional study design was conducted from February 23 to June 23, 2020 at Ethiopian public health. A structured questionnaire was used to collect data on socio-demographic factors and clinical conditions. Blood specimens were analyzed using standard microbiological techniques. Antimicrobial susceptibility tests were performed using Kirby-Bauer disc diffusion technique and Vitek compact 2. Simple and multiple logistic regressions were used to assess the potential risk factors.A total of 175 pathogens isolated from 346 blood specimens. Of these, 60% Gram-negative bacteria, 30.86% Gram-positive bacteria and 9.14% fungal isolates were identified. Burkholderia cepacia and Coagulase negative staphylococcus were the predominant pathogen among Gram-negative and Gram-positive bacteria respectively. Among fungus, Candida krusei (56.25%) was the most predominant isolate. The highest proportions of antibacterial resistance were observed among 3rd generation cephalosporin and penicillin. Most fungal isolates expressed resistance to fluconazole. Sex (P = 0.007), age (P < 0.001) and use of invasive medical devices (P = 0.003) were identified as risk factors for bacterial blood stream infections.The study showed high prevalence of blood stream infection was due to B. cepacia and non-C. albicans spp. This finding alarming ongoing investigation of blood stream infection is important for recognizing future potential preventive strategies including environmental hygiene and management of comorbid medical diseases to reduce the problem.© 2021. The Author(s).
Nosography of systemic inflammatory response syndrome, sepsis, severe sepsis, septic shock, and multiple organ dysfunction syndrome in internal medicine patients
Sepsis and nosocomial infection: patient characteristics, mechanisms, and modulation
DOI:10.3389/fimmu.2018.02446
PMID:30459764
[Cited within: 1]

Sepsis is a leading cause of death worldwide. After initial trials modulating the hyperinflammatory phase of sepsis failed, generations of researchers have focused on evaluating hypo-inflammatory immune phenotypes. The main goal has been to develop prognostic biomarkers and therapies to reduce organ dysfunction, nosocomial infection, and death. The depressed host defense in sepsis has been characterized by broad cellular reprogramming including lymphocyte exhaustion, apoptosis, and depressed cytokine responses. Despite major advances in this field, our understanding of the dynamics of the septic host response and the balance of inflammatory and anti-inflammatory cellular programs remains limited. This review aims to summarize the epidemiology of nosocomial infections and characteristic immune responses associated with sepsis, as well as immunostimulatory therapies currently under clinical investigation.