Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has posed a serious challenge to emergency departments that usually encounter emergencies and severe diseases.[1⇓⇓⇓-5] Angiotensin-converting enzyme 2 (ACE2), a protein that interacts with the viral spike protein(s), allows SARS-CoV-2 to penetrate epithelial cells.[6] There is mounting evidence that suggests that the digestive system may also be affected.[7] An observational study described potential patterns of pancreatic injury (elevated amylase and lipase) in patients with coronavirus disease 2019 (COVID-19).[8] Additionally, sporadic case reports have described secondary pancreatitis in patients with SARS-CoV-2 infection and presented imaging evidence.[9] Moreover, a previous study showed that COVID-19 patients not only have a higher risk of developing acute pancreatitis (AP), but also have a significantly higher mortality than those without COVID-19.[10] Using bioinformatics analysis, it may be possible to reveal how COVID-19 and AP are related. In this study, two RNA-seq datasets of SARS-CoV-2 and AP were selected for analysis.
METHODS
The microarray datasets of COVID-19 (GSE152641) and AP (GSE194331) samples were both obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/)to detect mutual differentially expressed genes (DEGs) (|log2FoldChange| ≥1.5 and adjusted P-value<0.05). A thorough investigation of the gene enrichment analysis was carried out to identify the functional enrichment of common DEGs. Then, a protein-protein interaction (PPI) network was constructed to extract the hub genes. In addition, the infiltration of various immune cells in the two diseases was analyzed based on the machine learning tool, Cell-type Identification By Estimating Relative Subsets of RNA Transcripts x (CIBERSORTx). The lncRNA-miRNA-mRNA network was constructed through the Encyclopedia of RNA Interactomes (ENCORI) database (http://starbase.sysu.edu.cn/).
RESULTS
Identification of common DEGs between COVID-19 and AP
Through analysis of GSE152641, we identified 1,216 DEGs and confirmed 399 DEGs from GSE194331. Volcano plots displayed dysregulated COVID-19 and AP genes, while comparison revealed 95 common DEGs (supplementary Figure 1).
Enrichment analysis of DEGs
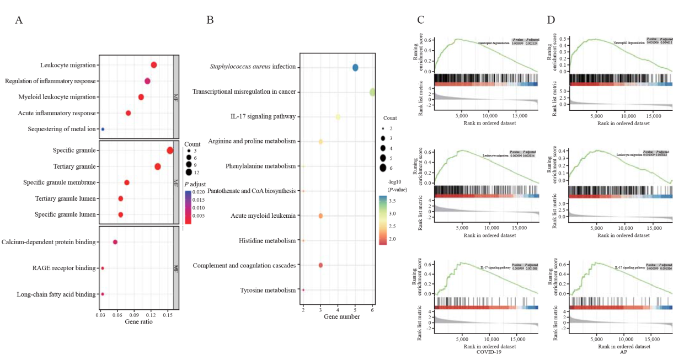
The top 10 items in the categories of molecular function (MF), biological process (BP), and cellular component (CC) of gene ontology (GO) are summarized in Figure 1A. In the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, the common DEGs were strongly involved in “Staphylococcus aureus infection”, “transcriptional misregulation in cancer” and “interleukin-17 (IL-17) signaling pathway” (Figure 1B). Similarly, gene set enrichment analysis (GSEA) revealed significant enrichment of immune-related pathways in both diseases (Figures 1 C and D). These results further suggest that similar immune responses may occur during the course of COVID-19 and AP.
Figure 1.
Figure 1.
Enrichment analyses of DEGs. A and B: results of GO and KEGG enrichment analysis; C and D: results of GSEA in COVID-19 and AP. RAGE: receptor for advanced glycation endproducts; GSEA: gene set enrichment analysis; DEGs: differentially expressed genes; GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; AP: acute pancreatitis.
PPI network construction and hub gene identification
The PPI network of common DEGs was constructed by string (supplementary Figure 2). The 20 hub genes with the highest scores were then determined using five algorithms based on Cytoscape I. Afterward, we confirmed 14 hub genes, which were all highly expressed in both diseases. Finally, a total of 20 predicted co-expressed genes were included in this co-expression pattern based on the multiple properties of the relationship. Among the 20 co-expressed genes, cathelicidin antimicrobial peptide (CAMP), folate receptor 3 (FOLR3), and interleukin-1 alpha (IL-1α) were included. Thus, they may be involved in the coordinated pathways that cause AP in response to COVID-19.
Evaluation and analysis of immune cell infiltration
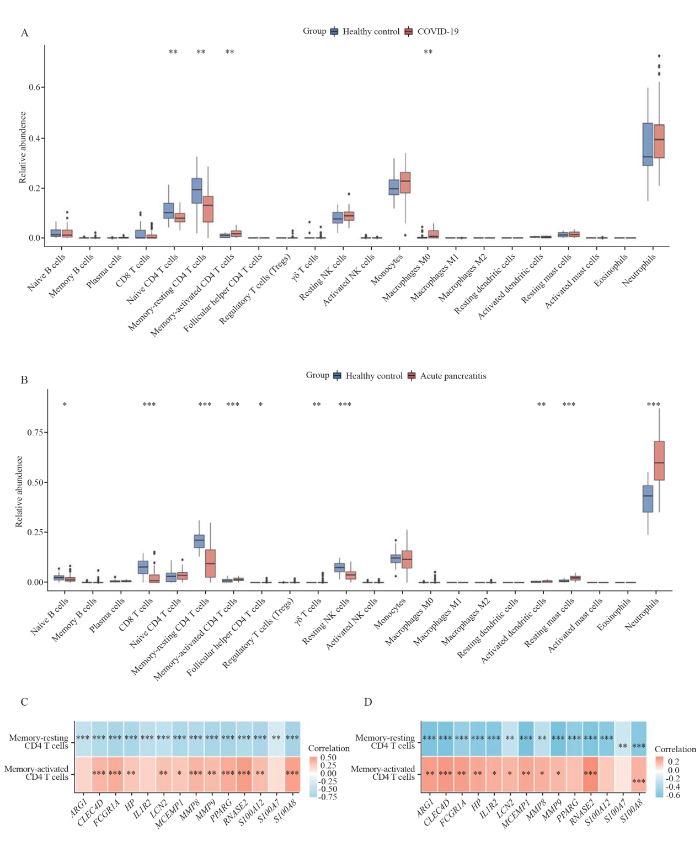
Considering that the enrichment analysis identified multiple immune-related pathways, we evaluated immune cell infiltration in both diseases (Figures 2 A and B). Among them, only memory-resting CD4 T cells and memory-activated CD4 T cells were significantly different in COVID-19 and AP at the same time, and the trend of difference between the two diseases was consistent. We analyzed the correlation between 14 hub genes and common infiltrating immune cells. Figures 2 C and D show that almost 14 hub genes were negatively correlated with memory-resting CD4 T cells and positively correlated with memory-activated CD4 T cells in both diseases.
Figure 2.
Figure 2.
Landscape of innate immune cell infiltration in COVID-19 and AP. A: landscape of innate immune cell infiltration in COVID-19. B: landscape of innate immune cell infiltration in AP. C: correlation of 14 hub genes with infiltrated immune cells in COVID-19. D: correlation of 14 hub genes with infiltrated immune cells in AP. A red box indicates a positive correlation, blue indicates a negative correlation, and P-values were corrected for false discovery rate. *P<0.05, **P<0.01, ***P<0.001.
Construction of transcriptional regulatory networks
We aimed to detect substantial changes occurring at the transcriptional level and gain insights into the hub genes (supplementary Figure 3). The results showed that 49 transcription factor (TF) targets were predicted from the Jellyfish-based Assembly Sequence Polisher for Error Reduction (JASPER) database to generate the co-regulatory network (63 nodes and 101 edges). Two critical genes (PPARG and S100A8) had a higher degree. In addition, a miRNA-gene interaction network was built. The hsa-miR-27a-3p was highly enriched.
LncRNA-miRNA-mRNA ceRNA regulatory network
The lncRNA-miRNA-mRNA ceRNA regulatory network can lead to a much better understanding of gene function and regulation. The ceRNA regulatory network was visualized using Cytoscape and included 64 lncRNAs, 77 miRNAs and 14 mRNAs (supplementary Figure 4).
DISCUSSION
The exact relationship between COVID-19 and AP, however, is still controversial and difficult to understand. The following is a summary of the explanations that could explain the relationship between AP and COVID-19: (1) Since ACE2 receptor proteins are present in the pancreatic ductal, acinar, and islet cells, infection of the pancreas with SARS-CoV-2 is undoubtedly possible.[11] (2) Alterations in inflammatory responses induced by SARS-CoV-2 infection may result in pancreatic damage.[12,13] (3) Additionally, medications used to treat COVID-19 may have a negative impact on the development of AP. To develop new tactics, it is vital to reveal the exact relationship between COVID-19 and AP. In this study, we used bioinformatics-related methods to investigate the potential interaction between COVID-19 and AP.
Inflammation may play a substantial role in the mechanism of COVID-19-induced AP according to the findings of BP term GO annotation analysis, which showed that the DEGs were considerably enriched in leukocyte migration and controlled inflammatory response. The notable GO-CC terms, such as tertiary granule, special granule, and secretory granule lumen, are related to neutrophil degranulation. Simultaneously, according to the enrichment pathway results, the common DEGs primarily focused on neutrophil degranulation and the IL-17 signaling pathway. The activated neutrophils play a critical role in the pathophysiology and progression of AP by initiating chemokine and cytokine cascades.[14] In addition, the IL-17 level and neutrophilia have been identified as markers associated with poor prognosis and severe respiratory symptoms in patients with COVID-19. Taken together, monitoring neutrophil activity may help to identify AP in COVID-19. In addition, MF analysis of GO terms showed that the DEGs were highly enriched for receptor for advanced glycation endproducts (RAGE) binding, calcium-dependent protein binding, and long-chain fatty acids binding. Leukocyte calcium-dependent binding proteins are essential for inflammatory cell transmigration into epithelial tissue. Previous research has demonstrated that calcium-binding protein increases the severity of pancreatitis and intra-acinar events.[15] On the other hand, RAGE may not only contribute to the tissue damage induced by SARS-CoV-2 infection,[16] but also contribute to the development of pancreatitis by mediating nucleosome-induced absent in melanoma 2 (AIM2) inflammasome activation and the release of proinflammatory mediators from macrophages.[17]
Both COVID-19 and AP have a pathophysiology that is strongly influenced by immune inflammation.[18] According to the results of immune infiltration, COVID-19 patients and AP patients showed decreased memory-resting CD4 T cells and increased memory-activated CD4 T cells. T-cell immunity plays an indispensable role in the control of primary SARS-CoV-2 infection.[19] CD4 T cells have the ability to differentiate into several different cell types with the capacity to have direct antiviral activities. Among them, T helper type 1 (Th1) cells provide direct antiviral functions via the production of interferon-gamma (IFN-γ).[20] In animal studies, IFN-γ has been shown to increase serum interleukin (IL)-18 and reduce serum IL-27 to aggravate inflammation in AP.[21] Therefore, the activation of memory CD4 T cells may be the cause of COVID-19-induced AP, which needs further verification and support by more experiments. We further deduced that hub genes highly expressed in both diseases, in particular ARG1, FCGR1A, LCN2, and MCEMP1, may cause the emergence of AP in patients with COVID-19. Accordingly, ARG1 has been demonstrated to be significantly upregulated in the whole blood of COVID-19 patients, suggesting that it may be a useful diagnostic biomarker.[22]
Non-coding RNAs (ncRNAs), including miRNAs and lncRNAs, cannot encode proteins, but they can perform their functions by regulating coding RNAs. The application of ncRNAs as biomarkers or intervention targets can provide new insights into the diagnosis and treatment of diseases. A coding transcriptional map of the peripheral immune response in COVID-19 patients revealed that hsa-miR-146a-5p is a crucial player in the etiology of the disease and acts as a hub regulator of the host immune response,[23] which is consistent with our findings. The hsa-miR-27a-3p, which was the most highly expressed miRNA in our study, has been identified as a crucial factor in regulating protein ubiquitination of SARS-CoV-2 infection.[24] We demonstrated the potential correlation between hsa-miR-34a-5p and AP, although further studies are needed. Our findings provide further details on the possible pathogenic mechanism behind COVID-19-induced AP and lay the groundwork for further research.
CONCLUSIONS
Our results provide evidence that the altered inflammatory responses induced by SARS-CoV-2 infection may be the cause of COVID-19-induced AP. The activation of memory CD4 T cells may be the main cause of COVID-19-induced AP. These findings provide information for those trying to understand the underlying relationships between COVID-19 and AP.
ACKNOWLEDGMENTS
We thank all the patients who participated in this study and donated samples, as well as the GEO database for providing their platform.
Funding: This study was sponsored by the Key Research and Development Program of Zhejiang Province (2019C03076). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Ethical approval: Not applicable.
Conflicts of interest: The authors declare that they have no conflicts of interest.
Contributors: ZDW and PW contributed equally to this work and share first authorship. ZDW and PW conceived, designed this research and drafted the manuscript. ZDW carried out the data analysis and data interpretation. PW is responsible for literature searching on the background and the image processing. XL and CYS annotated the picture and wrote the conclusion. JS and LL searched on the background of diseases and proofread the references. YQL reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
All the supplementary files in this paper are available at http://wjem.com.cn.
Reference
The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2
DOI:10.1038/s41564-020-0695-z
PMID:32123347
[Cited within: 1]

The present outbreak of a coronavirus-associated acute respiratory disease called coronavirus disease 19 (COVID-19) is the third documented spillover of an animal coronavirus to humans in only two decades that has resulted in a major epidemic. The Study Group (CSG) of the International Committee on Taxonomy of Viruses, which is responsible for developing the classification of viruses and taxon nomenclature of the family, has assessed the placement of the human pathogen, tentatively named 2019-nCoV, within the. Based on phylogeny, taxonomy and established practice, the CSG recognizes this virus as forming a sister clade to the prototype human and bat severe acute respiratory syndrome coronaviruses (SARS-CoVs) of the species, and designates it as SARS-CoV-2. In order to facilitate communication, the CSG proposes to use the following naming convention for individual isolates: SARS-CoV-2/host/location/isolate/date. While the full spectrum of clinical manifestations associated with SARS-CoV-2 infections in humans remains to be determined, the independent zoonotic transmission of SARS-CoV and SARS-CoV-2 highlights the need for studying viruses at the species level to complement research focused on individual pathogenic viruses of immediate significance. This will improve our understanding of virus–host interactions in an ever-changing environment and enhance our preparedness for future outbreaks.
Tripterygium wilfordii Hook.f. ameliorates paraquat-induced lung injury by reducing oxidative stress and ferroptosis via Nrf2/HO-1 pathway
DOI:10.1016/j.ecoenv.2023.114575 URL [Cited within: 1]
Isolated superior mesenteric artery rupture caused by abdominal trauma
DOI:10.1631/jzus.B2200288 [Cited within: 1]
Inflammatory granuloma of the trachea: a rare case with Epstin-Barr virus infection
DOI:10.1631/jzus.B2300024 [Cited within: 1]
Perforation of the esophagus: an overlooked cause of chest pain as a complication of esophageal foreign bodies
DOI:10.1631/jzus.B2300026 [Cited within: 1]
Polymorphisms in ACE, ACE2, AGTR1 genes and severity of COVID-19 disease
DOI:10.1371/journal.pone.0263140
URL
[Cited within: 1]

Infection by the SARS-Cov-2 virus produces in humans a disease of highly variable and unpredictable severity. The presence of frequent genetic single nucleotide polymorphisms (SNPs) in the population might lead to a greater susceptibility to infection or an exaggerated inflammatory response. SARS-CoV-2 requires the presence of the ACE2 protein to enter in the cell and ACE2 is a regulator of the renin-angiotensin system. Accordingly, we studied the associations between 8 SNPs from AGTR1, ACE2 and ACE genes and the severity of the disease produced by the SARS-Cov-2 virus.
Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China
DOI:S0140-6736(20)30183-5
PMID:31986264
[Cited within: 1]

A recent cluster of pneumonia cases in Wuhan, China, was caused by a novel betacoronavirus, the 2019 novel coronavirus (2019-nCoV). We report the epidemiological, clinical, laboratory, and radiological characteristics and treatment and clinical outcomes of these patients.All patients with suspected 2019-nCoV were admitted to a designated hospital in Wuhan. We prospectively collected and analysed data on patients with laboratory-confirmed 2019-nCoV infection by real-time RT-PCR and next-generation sequencing. Data were obtained with standardised data collection forms shared by WHO and the International Severe Acute Respiratory and Emerging Infection Consortium from electronic medical records. Researchers also directly communicated with patients or their families to ascertain epidemiological and symptom data. Outcomes were also compared between patients who had been admitted to the intensive care unit (ICU) and those who had not.By Jan 2, 2020, 41 admitted hospital patients had been identified as having laboratory-confirmed 2019-nCoV infection. Most of the infected patients were men (30 [73%] of 41); less than half had underlying diseases (13 [32%]), including diabetes (eight [20%]), hypertension (six [15%]), and cardiovascular disease (six [15%]). Median age was 49·0 years (IQR 41·0-58·0). 27 (66%) of 41 patients had been exposed to Huanan seafood market. One family cluster was found. Common symptoms at onset of illness were fever (40 [98%] of 41 patients), cough (31 [76%]), and myalgia or fatigue (18 [44%]); less common symptoms were sputum production (11 [28%] of 39), headache (three [8%] of 38), haemoptysis (two [5%] of 39), and diarrhoea (one [3%] of 38). Dyspnoea developed in 22 (55%) of 40 patients (median time from illness onset to dyspnoea 8·0 days [IQR 5·0-13·0]). 26 (63%) of 41 patients had lymphopenia. All 41 patients had pneumonia with abnormal findings on chest CT. Complications included acute respiratory distress syndrome (12 [29%]), RNAaemia (six [15%]), acute cardiac injury (five [12%]) and secondary infection (four [10%]). 13 (32%) patients were admitted to an ICU and six (15%) died. Compared with non-ICU patients, ICU patients had higher plasma levels of IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A, and TNFα.The 2019-nCoV infection caused clusters of severe respiratory illness similar to severe acute respiratory syndrome coronavirus and was associated with ICU admission and high mortality. Major gaps in our knowledge of the origin, epidemiology, duration of human transmission, and clinical spectrum of disease need fulfilment by future studies.Ministry of Science and Technology, Chinese Academy of Medical Sciences, National Natural Science Foundation of China, and Beijing Municipal Science and Technology Commission.Copyright © 2020 Elsevier Ltd. All rights reserved.
Pancreatic injury patterns in patients with coronavirus disease 19 pneumonia
DOI:S0016-5085(20)30409-1 PMID:32247022 [Cited within: 1]
SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas
DOI:10.1038/s42255-021-00347-1
PMID:33536639
[Cited within: 1]

Infection-related diabetes can arise as a result of virus-associated β-cell destruction. Clinical data suggest that the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing the coronavirus disease 2019 (COVID-19), impairs glucose homoeostasis, but experimental evidence that SARS-CoV-2 can infect pancreatic tissue has been lacking. In the present study, we show that SARS-CoV-2 infects cells of the human exocrine and endocrine pancreas ex vivo and in vivo. We demonstrate that human β-cells express viral entry proteins, and SARS-CoV-2 infects and replicates in cultured human islets. Infection is associated with morphological, transcriptional and functional changes, including reduced numbers of insulin-secretory granules in β-cells and impaired glucose-stimulated insulin secretion. In COVID-19 full-body postmortem examinations, we detected SARS-CoV-2 nucleocapsid protein in pancreatic exocrine cells, and in cells that stain positive for the β-cell marker NKX6.1 and are in close proximity to the islets of Langerhans in all four patients investigated. Our data identify the human pancreas as a target of SARS-CoV-2 infection and suggest that β-cell infection could contribute to the metabolic dysregulation observed in patients with COVID-19.
SARS-CoV-2 infection in acute pancreatitis increases disease severity and 30-day mortality: COVID PAN collaborative study
DOI:10.1136/gutjnl-2020-323364
PMID:33547182
[Cited within: 1]

There is emerging evidence that the pancreas may be a target organ of SARS-CoV-2 infection. This aim of this study was to investigate the outcome of patients with acute pancreatitis (AP) and coexistent SARS-CoV-2 infection.A prospective international multicentre cohort study including consecutive patients admitted with AP during the current pandemic was undertaken. Primary outcome measure was severity of AP. Secondary outcome measures were aetiology of AP, intensive care unit (ICU) admission, length of hospital stay, local complications, acute respiratory distress syndrome (ARDS), persistent organ failure and 30-day mortality. Multilevel logistic regression was used to compare the two groups.1777 patients with AP were included during the study period from 1 March to 23 July 2020. 149 patients (8.3%) had concomitant SARS-CoV-2 infection. Overall, SARS-CoV-2-positive patients were older male patients and more likely to develop severe AP and ARDS (p<0.001). Unadjusted analysis showed that SARS-CoV-2-positive patients with AP were more likely to require ICU admission (OR 5.21, p<0.001), local complications (OR 2.91, p<0.001), persistent organ failure (OR 7.32, p<0.001), prolonged hospital stay (OR 1.89, p<0.001) and a higher 30-day mortality (OR 6.56, p<0.001). Adjusted analysis showed length of stay (OR 1.32, p<0.001), persistent organ failure (OR 2.77, p<0.003) and 30-day mortality (OR 2.41, p<0.04) were significantly higher in SARS-CoV-2 co-infection.Patients with AP and coexistent SARS-CoV-2 infection are at increased risk of severe AP, worse clinical outcomes, prolonged length of hospital stay and high 30-day mortality.© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
COVID-19 and acute pancreatitis: examining the causality
DOI:10.1038/s41575-020-00389-y PMID:33203968 [Cited within: 1]
Immune dysfunction following COVID-19, especially in severe patients
DOI:10.1038/s41598-020-72718-9
[Cited within: 1]

The coronavirus disease 2019 (COVID-19) has been spreading worldwide. Severe cases quickly progressed with unfavorable outcomes. We aim to investigate the clinical features of COVID-19 and identify the risk factors associated with its progression. Data of confirmed SARS-CoV-2-infected patients and healthy participants were collected. Thirty-seven healthy people and 79 confirmed patients, which include 48 severe patients and 31 mild patients, were recruited. COVID-19 patients presented with dysregulated immune response (decreased T, B, and NK cells and increased inflammatory cytokines). Also, they were found to have increased levels of white blood cell, neutrophil count, and D-dimer in severe cases. Moreover, lymphocyte, CD4+ T cell, CD8+ T cell, NK cell, and B cell counts were lower in the severe group. Multivariate logistic regression analysis showed that CD4+ cell count, neutrophil-to-lymphocyte ratio (NLR) and D-dimer were risk factors for severe cases. Both CT score and clinical pulmonary infection score (CPIS) were associated with disease severity. The receiver operating characteristic (ROC) curve analysis has shown that all these parameters and scores had quite a high predictive value. Immune dysfunction plays critical roles in disease progression. Early and constant surveillance of complete blood cell count, T lymphocyte subsets, coagulation function, CT scan and CPIS was recommended for early screening of severe cases.
Comparison of clinical and immunological profiles in coronavirus disease 2019 and influenza patients: a case control study
DOI:10.5847/wjem.j.1920-8642.2022.042 URL [Cited within: 1]
NLRP3 inflammasome regulates development of systemic inflammatory response and compensatory anti-inflammatory response syndromes in mice with acute pancreatitis
DOI:S0016-5085(19)41413-3
PMID:31593700
[Cited within: 1]

Pancreatitis starts with primarily sterile local inflammation that induces systemic inflammatory response syndrome, followed by compensatory anti-inflammatory response syndrome (CARS). We investigated the mechanisms of these processes in mice and human serum.We induced severe acute pancreatitis by partial duct ligation with caerulein stimulation or intraperitoneal injection of l-arginine in mice with deletion of interleukin (IL)12B, NLRP3, or IL18 and in mice given MCC950, a small molecule inhibitor of the NLRP3-inflammasome. Pancreata were collected from mice and analyzed by histology, and cytokine levels were measured in serum samples. We measured activation of adaptive immune responses in mice with pancreatitis by flow cytometry analysis of T cells (CD25 and CD69) isolated from the spleen. Differentiation of T-helper (Th1) cells, Th2 cells, and T-regulatory cells was determined by nuclear staining for TBET, GATA3, and FOXP3. We performed transcriptome analysis of mouse lymph nodes and bone marrow-derived macrophages after incubation with acini. We measured levels of cytokines in serum samples from patients with mild and severe acute pancreatitis.Activation of the adaptive immune response in mice was initiated by macrophage-derived, caspase 1-processed cytokines and required activation of NLRP3 (confirmed in serum samples from patients with pancreatitis). Spleen cells from mice with pancreatitis had increases in Th2 cells but not in Th1 cells. Bone marrow-derived macrophages secreted IL1B and IL18, but not IL12, after co-incubation with pancreatic acini. T-cell activation and severity of acute pancreatitis did not differ significantly between IL12B-deficient and control mice. In contrast, NLRP3- or IL18-deficient mice had reduced activation of T cells and no increase in Th2 cell-mediated responses compared with control mice. The systemic type 2 immune response was mediated by macrophage-derived cytokines of the IL1 family. Specifically, IL18 induced a Th2 cell-mediated response in the absence of IL12. MCC950 significantly reduced neutrophil infiltration, T-cell activation, and disease severity in mice.In mice with severe pancreatitis, we found systemic inflammatory response syndrome and compensatory anti-inflammatory response syndrome developed in parallel. Infiltrating macrophages promote inflammation and simultaneously induce a Th2 cell-mediated response via IL18. Inhibition of NLRP3 reduces systemic inflammatory response syndrome and compensatory anti-inflammatory response syndrome and might be used to treat patients with severe pancreatitis.Copyright © 2020 AGA Institute. Published by Elsevier Inc. All rights reserved.
Pancreatic ductal deletion of S100A9 alleviates acute pancreatitis by targeting VNN1-mediated ROS release to inhibit NLRP3 activation
DOI:10.7150/thno.54245
PMID:33754072
[Cited within: 1]

Recent studies have proven that the overall pathophysiology of pancreatitis involves not only the pancreatic acinar cells but also duct cells, however, pancreatic duct contribution in acinar cells homeostasis is poorly known and the molecular mechanisms leading to acinar insult and acute pancreatitis (AP) are unclear. Our previous work also showed that S100A9 protein level was notably increased in AP rat pancreas through iTRAQ-based quantitative proteomic analysis. Therefore, we investigated the actions of injured duct cells on acinar cells and the S100A9-related effects and mechanisms underlying AP pathology in the present paper. In this study, we constructed S100A9 knockout (s100a9) mice and an coculture system for pancreatic duct cells and acinar cells. Moreover, a variety of small molecular inhibitors of S100A9 were screened from ChemDiv through molecular docking and virtual screening methods. We found that the upregulation of S100A9 induces cell injury and inflammatory response via NLRP3 activation by targeting VNN1-mediated ROS release; and loss of S100A9 decreases AP injury and. Moreover, molecular docking and mutant plasmid experiments proved that S100A9 has a direct interaction with VNN1 through the salt bridges formation of Lys57 and Glu92 residues in S100A9 protein. We further found that compounds CHNO and CHFNOS can significantly improve AP injury and through inhibiting S100A9-VNN1 interaction. Our study showed the important regulatory effect of S100A9 on pancreatic duct injury during AP and revealed that inhibition of the S100A9-VNN1 interaction may be a key therapeutic target for this disease.© The author(s).
S100A8, S100A9 and S100A12 activate airway epithelial cells to produce MUC5AC via extracellular signal-regulated kinase and nuclear factor-κB pathways
DOI:10.1111/imm.12352
PMID:24975020
[Cited within: 1]

Airway mucus hyperproduction is a common feature of chronic airway diseases such as severe asthma, chronic obstructive pulmonary disease and cystic fibrosis, which are closely associated with neutrophilic airway inflammation. S100A8, S100A9 and S100A12 are highly abundant proteins released by neutrophils and have been identified as important biomarkers in many inflammatory diseases. Herein, we report a new role for S100A8, S100A9 and S100A12 for producing MUC5AC, a major mucin protein in the respiratory tract. All three S100 proteins induced MUC5AC mRNA and the protein in normal human bronchial epithelial cells as well as NCI-H292 lung carcinoma cells in a dose-dependent manner. A Toll-like receptor 4 (TLR4) inhibitor almost completely abolished MUC5AC expression by all three S100 proteins, while neutralization of the receptor for advanced glycation end-products (RAGE) inhibited only S100A12-mediated production of MUC5AC. The S100 protein-mediated production of MUC5AC was inhibited by the pharmacological agents that block prominent signalling molecules for MUC5AC expression, such as mitogen-activated protein kinases, nuclear factor-κB (NF-κB) and epidermal growth factor receptor. S100A8, S100A9 and S100A12 equally elicited both phosphorylation of extracellular signal-regulated kinase (ERK) and nuclear translocation of NF-κB/degradation of cytosolic IκB with similar kinetics through TLR4. In contrast, S100A12 preferentially activated the ERK pathway rather than the NF-κB pathway through RAGE. Collectively, these data reveal the capacity of these three S100 proteins to induce MUC5AC production in airway epithelial cells, suggesting that they all serve as key mediators linking neutrophil-dominant airway inflammation to mucin hyperproduction. © 2014 John Wiley & Sons Ltd.
The receptor for advanced glycation end products activates the AIM2 inflammasome in acute pancreatitis
DOI:10.4049/jimmunol.1502340
PMID:27045109
[Cited within: 1]

Severe acute pancreatitis (AP) is responsible for significant human morbidity and mortality worldwide. Currently, no specific treatments for AP exist, primarily due to the lack of a mechanistic understanding of sterile inflammation and the resultant multisystem organ dysfunction, the pathologic response of AP linked to early death. In this study, we demonstrate that the class III major histocompatibility region III receptor for advanced glycation end products (RAGE) contributes to AP by modulating inflammasome activation in macrophages. RAGE mediated nucleosome-induced absent in melanoma 2 (but not NLRP3) inflammasome activation by modulating dsRNA-dependent protein kinase phosphorylation in macrophages. Pharmacological and genetic inhibition of the RAGE-dsRNA-dependent protein kinase pathway attenuated the release of inflammasome-dependent exosomal leaderless cytokines (e.g., IL-1β and high-mobility group box 1) in vitro. RAGE or absent in melanoma 2 depletion in mice limited tissue injury, reduced systemic inflammation, and protected against AP induced by l-arginine or cerulein in experimental animal models. These findings define a novel role for RAGE in the propagation of the innate immune response with activation of the nucleosome-mediated inflammasome and will help guide future development of therapeutic strategies to treat AP.Copyright © 2016 by The American Association of Immunologists, Inc.
Cytokines and inflammatory mediators: markers involved in interstitial damage to the pancreas in two dengue fever cases associated with acute pancreatitis
DOI:10.1371/journal.pone.0262785
URL
[Cited within: 1]

Dengue viral (DENV) infections can lead to acute pancreatitis and associated tissue damage. This study examined the pancreas from two fatal cases of DENV for histopathological changes as well as for the detection of cytokines, and other inflammatory mediators. Tissue sections were prepared for examination by ultrastructural and histopathological techniques. Sections from the pancreas of non-infected individuals were prepared in parallel as a control. The presence of viral replication in macrophages was detected by co-staining for the proteins NS3 and CD68 by immunofluorescence. Immunohistochemistry was used to detect cells that expressed cytokines and inflammatory mediators to characterize the inflammatory response. Edema, acinar necrosis and fibrosis areas associated with a mononuclear infiltrate were found in infected tissues. The major site of virus replication appeared to be macrophages based on their exclusive presentation of the viral protein NS3. Pancreatic tissues from the infected individuals also displayed increased levels of high mobility group box-1, caspase-3, gelatinase B and tumor necrosis factor alpha compared to controls. The presence of virus replicating macrophages in the pancreas was associated with multiple changes in tissue structure that included elevated levels of cytokines and inflammatory markers that may differentiate acute pancreatitis due to DENV infections from other causes.
Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity
DOI:10.1016/j.cell.2020.09.038
PMID:33010815
[Cited within: 1]

Limited knowledge is available on the relationship between antigen-specific immune responses and COVID-19 disease severity. We completed a combined examination of all three branches of adaptive immunity at the level of SARS-CoV-2-specific CD4 and CD8 T cell and neutralizing antibody responses in acute and convalescent subjects. SARS-CoV-2-specific CD4 and CD8 T cells were each associated with milder disease. Coordinated SARS-CoV-2-specific adaptive immune responses were associated with milder disease, suggesting roles for both CD4 and CD8 T cells in protective immunity in COVID-19. Notably, coordination of SARS-CoV-2 antigen-specific responses was disrupted in individuals ≥ 65 years old. Scarcity of naive T cells was also associated with aging and poor disease outcomes. A parsimonious explanation is that coordinated CD4 T cell, CD8 T cell, and antibody responses are protective, but uncoordinated responses frequently fail to control disease, with a connection between aging and impaired adaptive immune responses to SARS-CoV-2.Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Adaptive immunity to SARS-CoV-2 and COVID-19
DOI:10.1016/j.cell.2021.01.007
PMID:33497610
[Cited within: 1]

The adaptive immune system is important for control of most viral infections. The three fundamental components of the adaptive immune system are B cells (the source of antibodies), CD4 T cells, and CD8 T cells. The armamentarium of B cells, CD4 T cells, and CD8 T cells has differing roles in different viral infections and in vaccines, and thus it is critical to directly study adaptive immunity to SARS-CoV-2 to understand COVID-19. Knowledge is now available on relationships between antigen-specific immune responses and SARS-CoV-2 infection. Although more studies are needed, a picture has begun to emerge that reveals that CD4 T cells, CD8 T cells, and neutralizing antibodies all contribute to control of SARS-CoV-2 in both non-hospitalized and hospitalized cases of COVID-19. The specific functions and kinetics of these adaptive immune responses are discussed, as well as their interplay with innate immunity and implications for COVID-19 vaccines and immune memory against re-infection.Copyright © 2021 Elsevier Inc. All rights reserved.
Effect of interferon-γ on NF-κB and cytokine IL-18 and IL-27 in acute pancreatitis
DOI:10.17305/bjbms.2013.2391 URL [Cited within: 1]
Arginase 1 (Arg1) as an up-regulated gene in COVID-19 patients: a promising marker in COVID-19 immunopathy
The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19
DOI:10.1002/ctm2.200
PMID:33135345
[Cited within: 1]

COVID-19 is currently a global pandemic, but the response of human immune system to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection remains unclear. Noncoding RNAs serve as immune regulators and thus may play a critical role in disease progression.We performed multi-transcriptome sequencing of both noncoding RNAs and mRNAs isolated from the red blood cell depleted whole blood of moderate and severe COVID-19 patients. The functions of noncoding RNAs were validated by analyses of the expression of downstream mRNAs. We further utilized the single-cell RNA-seq data of COVID-19 patients from Wilk et al. and Chua et al. to characterize noncoding RNA functions in different cell types.We defined four types of microRNAs with different expression tendencies that could serve as biomarkers for COVID-19 progress. We also identified miR-146a-5p, miR-21-5p, miR-142-3p, and miR-15b-5p as potential contributors to the disease pathogenesis, possibly serving as biomarkers of severe COVID-19 and as candidate therapeutic targets. In addition, the transcriptome profiles consistently suggested hyperactivation of the immune response, loss of T-cell function, and immune dysregulation in severe patients.Collectively, these findings provide a comprehensive view of the noncoding and coding transcriptional landscape of peripheral immune cells during COVID-19, furthering our understanding and offering novel insights into COVID-19 pathogenesis.© 2020 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections
DOI:10.1016/j.isci.2020.101947 URL [Cited within: 1]